Nous présentons ici un coup d’œil approfondi aux travaux de science ouverte effectués par nos boursiers et boursières PCNO. Cette page met en vedette certaines de leurs réalisations et se concentre sur leur engagement envers le partage des données et leurs contributions au Portail de Données et au site de NeuroLibre.org.
Abhijit Chinchani
CONP scholar and University of British Columbia graduate student Abhijit Chinchani has published a peer review manuscript in the journal Cortex. An electrical engineer by training, Abhijit is now using his engineering skills to build analysis frameworks for large and complex neuroscience datasets.
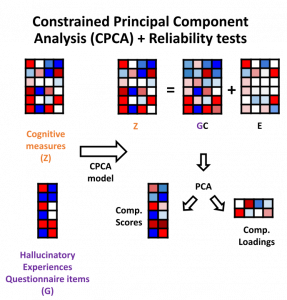
Traditional analyses can often oversimplify datasets by using weighted summary score methods which do not capture the richness of the data or the amount of variance for different metrics. In this most recent publication, titled “Item-specific overlap between hallucinatory experiences and cognition in the general population: A three-step multivariate analysis of international multi-site data”, Abhijit and the other scientists in the team took a brand new approach to look for links between psychiatric and other neurological symptoms.

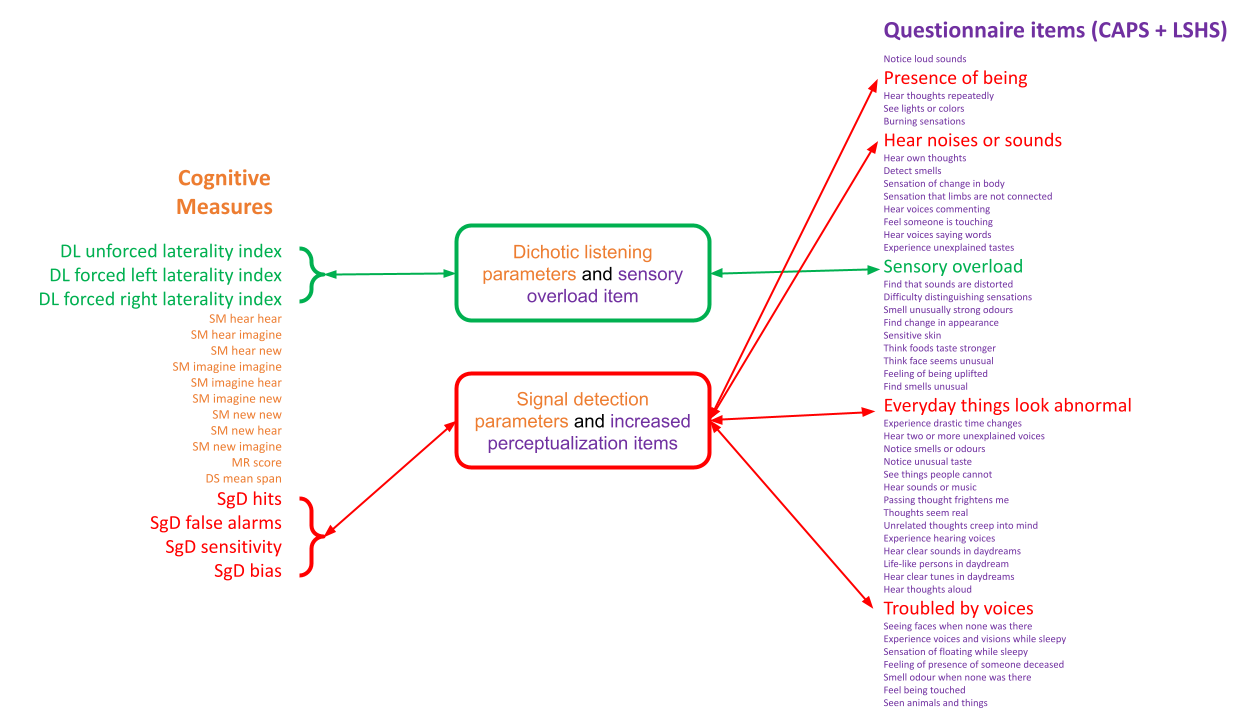
All of the findings of this exploratory study will soon be accessed openly through the CONP Portal. By making the code and the analysis framework publicly available, Abhijit hopes that other researchers will replicate his findings and perhaps apply this methodology to other datasets. Open science is an important part of the publication process for Abhijit, who believes transparency is critical in methods development: “it’s good for everyone and it’s good for the field”.
 You can learn more about Abhijit’s methods and data through his OSF archive and read his full manuscript in the journal Cortex.
You can learn more about Abhijit’s methods and data through his OSF archive and read his full manuscript in the journal Cortex.
Acknowledgements: The novel analysis method was developed in Dr. Todd Woodward’s Cognitive Neuroscience of Schizophrenia (CNOS) lab in collaboration with Dr. Heunsun Hwang (McGill University). Several members of the CNOS lab were involved in developing and testing the algorithm. Jeffrey LeDue, Dr. Tim Murphy, and other members of the dynamic brain circuits cluster helped in making the project open source. Mahesh Menon, Meighen Roes, Heungsun Hwang, Paul Allen, Vaughan Bell, Josef Bless, Catherine Bortolon, Matteo Cella, Charles Fernyhough, Jane Garrison, Eva Kozáková, Frank Larøi, Jamie Moffatt, Nicolas Say, Mimi Suzuki, Wei Lin Toh, Yuliya Zaytseva, Susan L. Rossell, Peter Moseley, and Todd S. Woodward were co-authors in the paper. The data used in this study was part of a larger international multisite study (https://journals.sagepub.com/doi/10.1177/0956797620985832). The data collection was supported by Wellcome Trust grant awarded to CF, NHMRC senior research fellowship to SLR, NHMRC New Investigator project grant to WLT, Internal Grant Agency of the Ministry of Health of the Czech Republic grant to EK and YZ, ERC Advanced Grant from the Helse-Vest Samarbeidsorganet and the Research Council of Norway to JB. To develop and test the novel method, AC was supported by Brain Canada, in partnership with Health Canada, for the Canadian Open Neuroscience Platform (CONP) initiative.
Mathieu Boudreau

Montreal Heart Institute research fellow and CONP Publication Committee member Mathieu Boudreau has written a chapter for a book called “Quantitative Magnetic Resonance Imaging”. Mathieu’s chapter – Quantitative T1 and T1ρ Mapping – describes measurement techniques for some of the fundamental parameters of Magnetic Resonance Imaging (MRI) and is available under the Creative Commons licence. In line with the principles of Open Science by which Mathieu conducts his research, the chapter was first published as a “chapter preprint” prior to peer review, similar to how researchers commonly share early versions of manuscripts on preprint servers before formal publication. Mathieu’s chapter can be found on the qMRLab blog and through NeuroLibre under an Creative Commons License.
Mathieu wanted his chapter to include interactive components and reproducible figures so that readers could interact with them in real time. Readers can alter parameters and re-execute the figures to see how outcomes are changed and can even edit and extract the code used to create these figures. The move away from static, flat figures that punctuate traditional research works is driven by the increasing need for reproducibility in research. Mathieu believes that laying bare how figures are generated is important for good science; “it offers transparency in terms of how we created these images”. The interactive elements of this chapter preprint are also the focus of Mathieu’s work on the NeuroLibre platform – a curated repository of interactive neuroscience notebooks that integrates data, text, code, and figures.
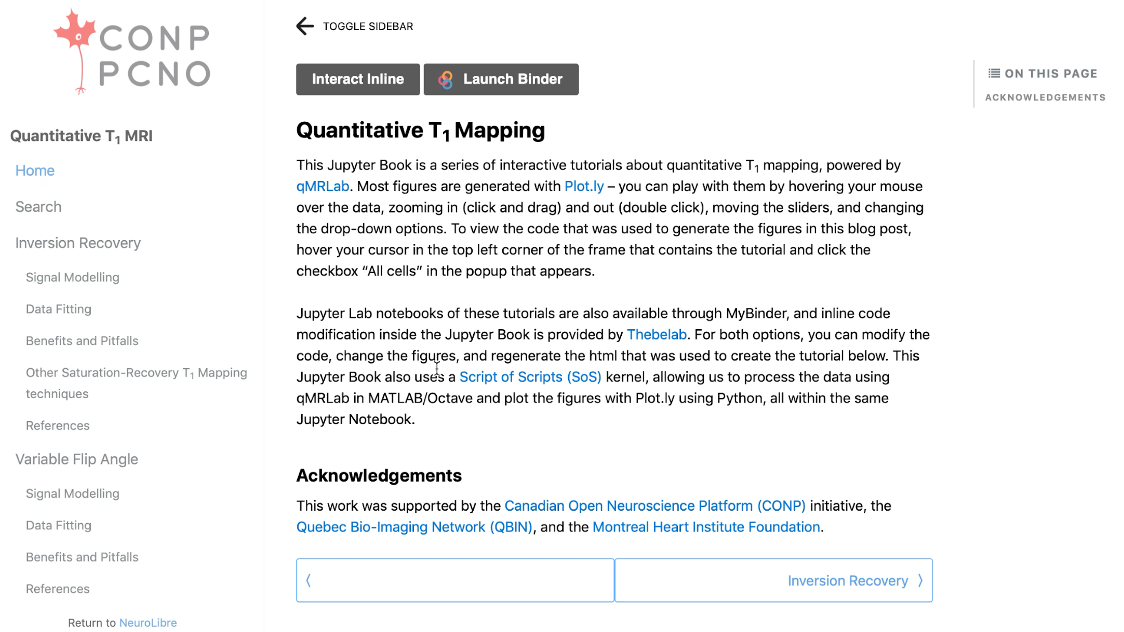
A further benefit of making the chapter available through the NeuroLibre platform is that users and readers of works archived there do not need to install any specialist or expensive software. Instead, everything may be run and accessed through a browser window. Taken in combination with the Open licenses by which all aspects of this work may be accessed, Mathieu believes this really helps with “accessibility of knowledge” by “removing barriers” for readers. Barriers and a lack of equitable access are ever-present issues in the field of MRI where over 90% of all research is conducted in the northern hemisphere. Mathieu’s chapter serves as an interactive tutorial, enabled by the NeuroLibre platform, to help make MRI technology more widely available to the global community.
More information about NeuroLibre can be found on their website.
Acknowledgements: Kathryn E. Keenan and Nikola Stikov are co-authors of this chapter. Agâh Karakuzu and Tommy Boshkovsky helped to create the framework for the interactive tutorial. The NeuroLibre team also includes Pierre Bellec and Jean-Baptiste Poline. This work was funded and supported by the Canadian Open Neuroscience Platform (CONP) initiative, the Quebec Bio-Imaging Network (QBIN), and the Montreal Heart Institute Foundation.
Jessica Royer

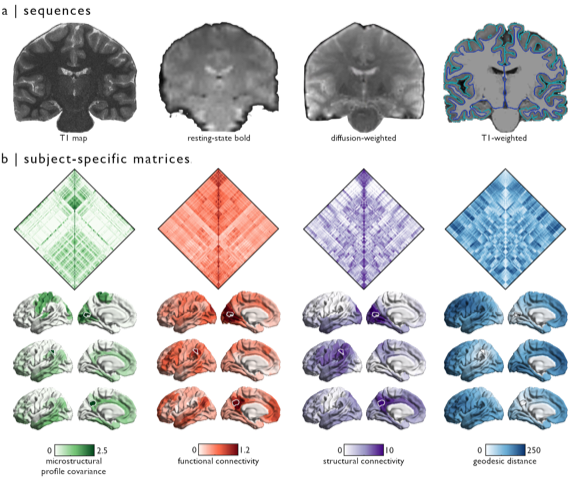
Ph.D. candidate and CONP Scholar Jessica Royer has contributed a new neuroimaging dataset to the CONP portal. Comprising data from 50 healthy human participants, this open dataset provides a wealth of information spanning anatomical structural sequence data, resting state functional acquisition and diffusion MRI.
These data, acquired in healthy control participants, will not only be a valuable resource for basic questions regarding brain structure and function but will also serve as a good comparator for similar datasets collected in patients with different brain diseases. Jessica is now working on an analogous dataset of patients with forms of epilepsy that do not respond to anti-seizure medication. She hopes to identify differences in brain organisation between the patient and control group, which might provide insight into these forms of epilepsy and lead to better treatments for patients.

Collection and curation of the MICA-MICs dataset has taken 3 years, with the first brain-imaging experiments beginning in 2018. Now, both the raw and processed data are available under open license through the CONP portal and are described in further detail in a preprint that is currently submitted for publication. Processing of the data was based on a fully open pipeline, so that other scientists can replicate their work or adapt the analyses to help answer their own research questions. This pre-processing pipeline was co-developed by Jessica and can be used to process any BIDS-compatible dataset.
For Jessica, open scientific practises are central to the research process. For these large, complex and expensive experiments, sharing the data makes sense to Jessica: “Putting the data out there will help other groups that might not have the resources to collect such data” she says. Working at The Neuro (Montreal Neurological Institute-Hospital), her lab is uniquely positioned to do this research in drug-resistant epilepsy patients given The Neuro’s long-standing expertise in epilepsy, its state of the art facilities, and a large number of patient referrals. By not only sharing the data and tools but also carefully annotating and curating them, Jessica hopes to increase usability by other research teams as much as possible.
To learn more about sharing your own neuroscience research through CONP or how to access existing datasets, you can learn more from our FAQ.
Acknowledgements: Several trainees of the Multimodal Imaging and Connectome Analysis Lab (MICA Lab) led by Boris Bernhardt contributed to data collection, pre-processing, and quality control of this dataset, including Shahin Tavakol, Hans Auer, Raul Rodriguez-Cruces, Sara Larivière, Reinder Vos de Wael, Oualid Benkarim, Casey Paquola, and Bo-yong Park. The MICA lab would also like to also highlight the help of Peer Herholz for his contribution to our pre-processing pipeline. This work was supported the Canadian Institute of Health Research (CIHR), Canadian Open Neuroscience Platform, the Canada Research Chairs (CRC) program, and National Sciences and Engineering Research Council of Canada (NSERC).
